多梳家族蛋白
多梳家族蛋白(英語:Polycomb-group proteins)是在果蠅中首次發現的能夠重塑染色質從而使基因在表觀遺傳上沉默的一類蛋白質家族。多梳家族蛋白因其可在黑腹果蠅的胚胎發育過程中通過調整染色質結構沉默同源異形基因而著名[1]。
昆蟲中
[編輯]果蠅屬的三胸家族(trxG)與多梳家族(PcG)蛋白的作用是相互拮抗的,他們都與被稱為細胞記憶模塊(CMMs)的染色體元件發生相互作用。三胸家族(trxG)蛋白維持着基因表達的活性狀態,多梳家族(PcG)蛋白通過抑制染色體活性而對沖這種激活作用,這種抑制功能在多個細胞世代中一直保持穩定,只能通過生殖細胞分化過程才能得到逆轉。多梳基因複合物所介導的沉默至少需要三類多蛋白複合物:PRC1(多梳抑制複合物1)、PRC2與PhoRC。這些複合物共同作用才能達到抑制的效果。
哺乳動物中
[編輯]多梳家族的基因表達在哺乳動物中對發育的多個方面有着重要作用[2]。Bmi1多梳RING指結構域蛋白促使神經幹細胞自我更新[3][4]。鼠科PRC2基因的無效突變體會導致胚胎死亡,但大多數PRC1的突變體則生下來就是同源異形突變體,後者導致圍生期死亡。相反的是PcG蛋白過表達會與多種腫瘤的嚴重程度和侵襲力成正相關[5]。哺乳動物的PRC1核心複合物與果蠅的非常相似。已知多梳Bmi1可調ink4基因座(p16Ink4a和p19Arf)[3][6]。
植物中
[編輯]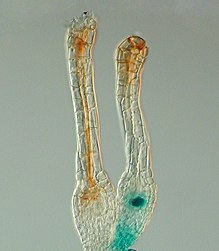
PcG蛋白FIE在小立碗蘚中特異性表達於如未受精卵細胞等的幹細胞中,這一點通過組織化學GUS染色後顯示出的藍色進行識別。在受精後的早期胚胎FIE基因很快失活[7][8]。
對於哺乳動物中:保持細胞處於分化狀態來說,PcG是必不可少的;植物中則不是這樣的:繼發地缺失PcG會導致去分化,促進胚胎發育[9]。
多梳家族蛋白通過沉默開花基因座C基因而干擾其對開花的控制[10]。開花基因座C基因是植物中抑制開花通路中的關鍵環節,在冬季該基因是被沉默的,這被認為是干擾植物春化現象的主要因素[11]。
參考文獻
[編輯]- ^ Portoso M, Cavalli G. The Role of RNAi and Noncoding RNAs in Polycomb Mediated Control of Gene Expression and Genomic Programming. Morris KV (編). RNA and the Regulation of Gene Expression: A Hidden Layer of Complexity. Caister Academic Press. 2008: 29–44 [2013-11-03]. ISBN 978-1-904455-25-7. (原始內容存檔於2014-01-02).
- ^ Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, Adli M, Kasif S, Ptaszek LM, Cowan CA, Lander ES, Koseki H, Bernstein BE. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLOS Genetics. 2008, 4 (10): e1000242 [2013-11-03]. PMID 18974828. doi:10.1371/journal.pgen.1000242. (原始內容存檔於2015-01-11).
- ^ 3.0 3.1 Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes & Development. 2005, 19 (12): 1432–1437 [2013-11-03]. PMC 1151659
 . PMID 15964994. doi:10.1101/gad.1299505. (原始內容存檔於2021-05-25).
. PMID 15964994. doi:10.1101/gad.1299505. (原始內容存檔於2021-05-25).
- ^ Park IK, Morrison SJ, Clarke MF. Bmi1, stem cells, and senescence regulation. Journal of Clinical Investigation. 2004, 113 (2): 175–179 [2013-11-03]. PMC 311443
 . PMID 14722607. doi:10.1172/JCI20800. (原始內容存檔於2020-07-28).
. PMID 14722607. doi:10.1172/JCI20800. (原始內容存檔於2020-07-28).
- ^ Sauvageau M, Sauvageau G. Polycomb group genes: keeping stem cell activity in balance. PLoS Biol. April 2008, 6 (4): e113. PMC 2689701
 . PMID 18447587. doi:10.1371/journal.pbio.0060113.
. PMID 18447587. doi:10.1371/journal.pbio.0060113.
- ^ Popov N, Gil J. Epigenetic regulation of the INK4b-ARF-INK4a locus: in sickness and in health (PDF). EPIGENETICS. 2010, 5 (8): 685–690 [2013-11-03]. PMC 3052884
 . PMID 20716961. doi:10.4161/epi.5.8.12996. (原始內容存檔於2020-05-11).
. PMID 20716961. doi:10.4161/epi.5.8.12996. (原始內容存檔於2020-05-11).
- ^ Mosquna A, Katz A, Decker EL, Rensing SA, Reski R, Ohad N. Regulation of stem cell maintenance by the Polyclmb protein FIE has been conserved during land plant evolution. Development. July 2009, 136 (14): 2433–44. PMID 19542356. doi:10.1242/dev.035048.
- ^ The Polycomb gene FIE is expressed (blue) in unfertilised egg cells of the moss Physcomitrella patens (right) and expression ceases after fertilisation in the developing diploid sporophyte (left). In situ GUS staining of two female sex organs (archegonia) of a transgenic plant expressing a translational fusion of FIE-uidA under control of the native FIE promoter. 存档副本. [2009-07-03]. (原始內容存檔於2009-06-26).
- ^ Aichinger E, Villar CB, Farrona S, Reyes JC, Hennig L, Köhler C. CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet. August 2009, 5 (8): e1000605. PMC 2718830
 . PMID 19680533. doi:10.1371/journal.pgen.1000605.
. PMID 19680533. doi:10.1371/journal.pgen.1000605.
- ^ Jiang D, Wang Y, Wang Y, He Y. Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS ONE. 2008, 3 (10): e3404. PMC 2561057
 . PMID 18852898. doi:10.1371/journal.pone.0003404.
. PMID 18852898. doi:10.1371/journal.pone.0003404.
- ^ Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES. The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. U.S.A. March 2000, 97 (7): 3753–8. PMC 16312
 . PMID 10716723. doi:10.1073/pnas.060023597.
. PMID 10716723. doi:10.1073/pnas.060023597.
深入閱讀
[編輯]- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. January 2007, 8 (1): 9–22. PMID 17173055. doi:10.1038/nrg1981.
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. February 2007, 128 (4): 735–45. PMID 17320510. doi:10.1016/j.cell.2007.02.009.
- Pirrotta V, Li HB. "A view of nuclear Polycomb bodies." Curr Opin Genet Dev. 2012 Apr;22(2):101-9. doi: 10.1016/j.gde.2011.11.004. PMID: 22178420 (頁面存檔備份,存於網際網路檔案館)
外部連結
[編輯]- polycomb group proteins. Humpath.com. (原始內容存檔於2020-09-30).
- The Polycomb and Trithorax page of the Cavalli lab (頁面存檔備份,存於網際網路檔案館) This page contains useful information on Polycomb and trithorax proteins, in the form of an introduction, links to published reviews, list of Polycomb and trithorax proteins, illustrative power point slides and a link to a genome browser showing the genome-wide distribution of these proteins in Drosophila melanogaster.
- Drosophila Genes in Development: Polycomb-group in the Homeobox Genes DataBase
- Chromatin organization and the Polycomb and Trithorax groups (頁面存檔備份,存於網際網路檔案館) in The Interactive Fly
